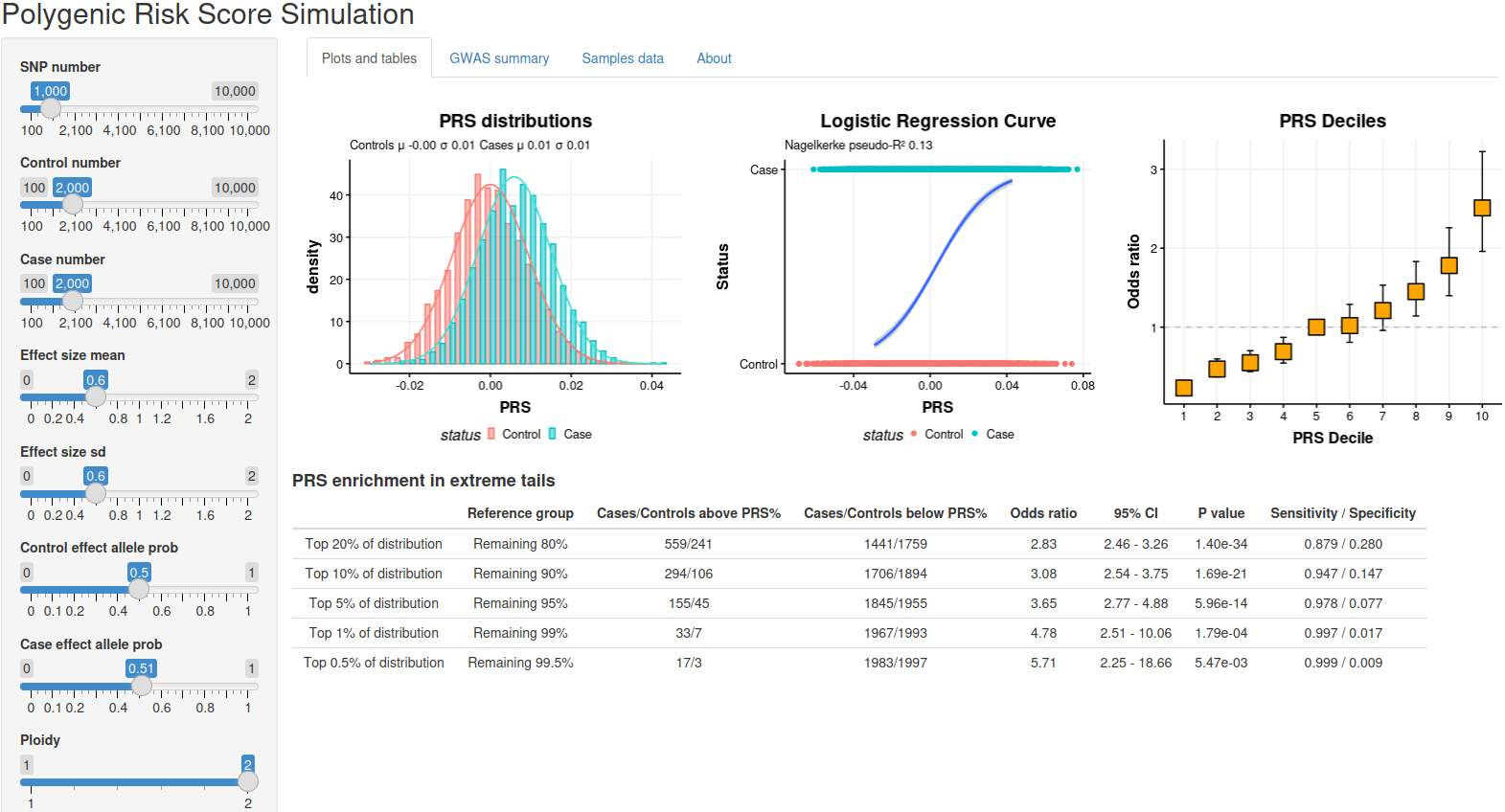
Brainspan gene plot
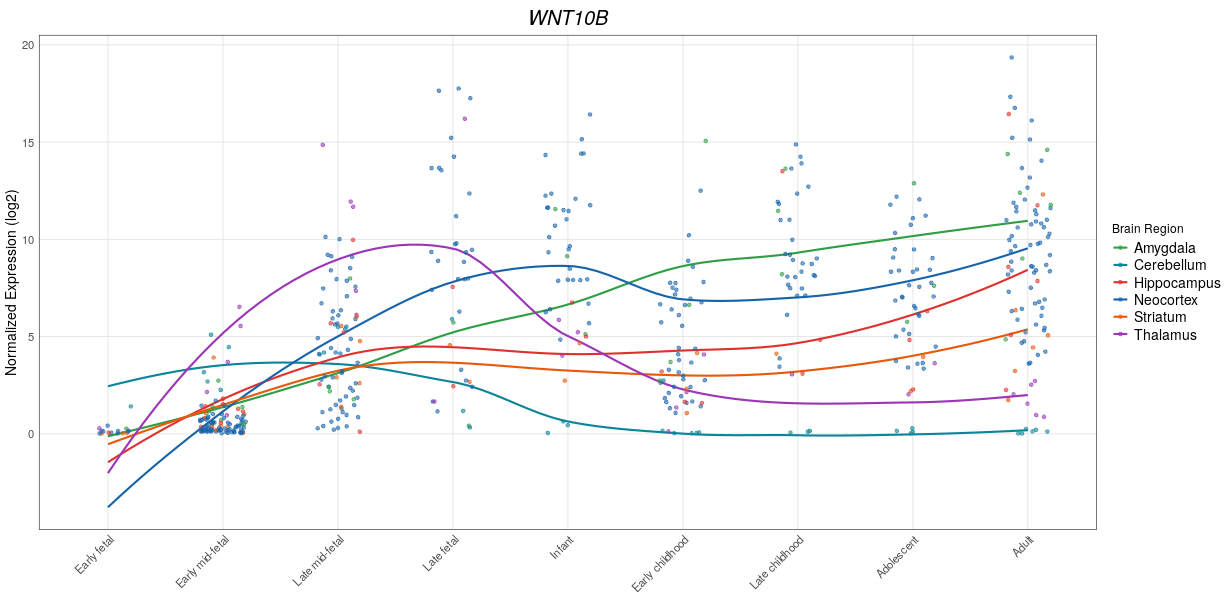
This tool plots expression the given gene in various brain structures in various developmental stages and ages. Image is available at dockerhub.
Var2TexShade

This tools visualizes homology of given protein change e.g. NP_001501.2:p.Arg121Leu using texshade. Texshade is a latex package for setting nucleotide and peptide alignments. Multiple sequence alignments for the proteins are retrieved from NCBI homologene database. The tool is useful for visualizing a variant on evulotionary conserved regions. The source code is available on GitHub.
HGNC version
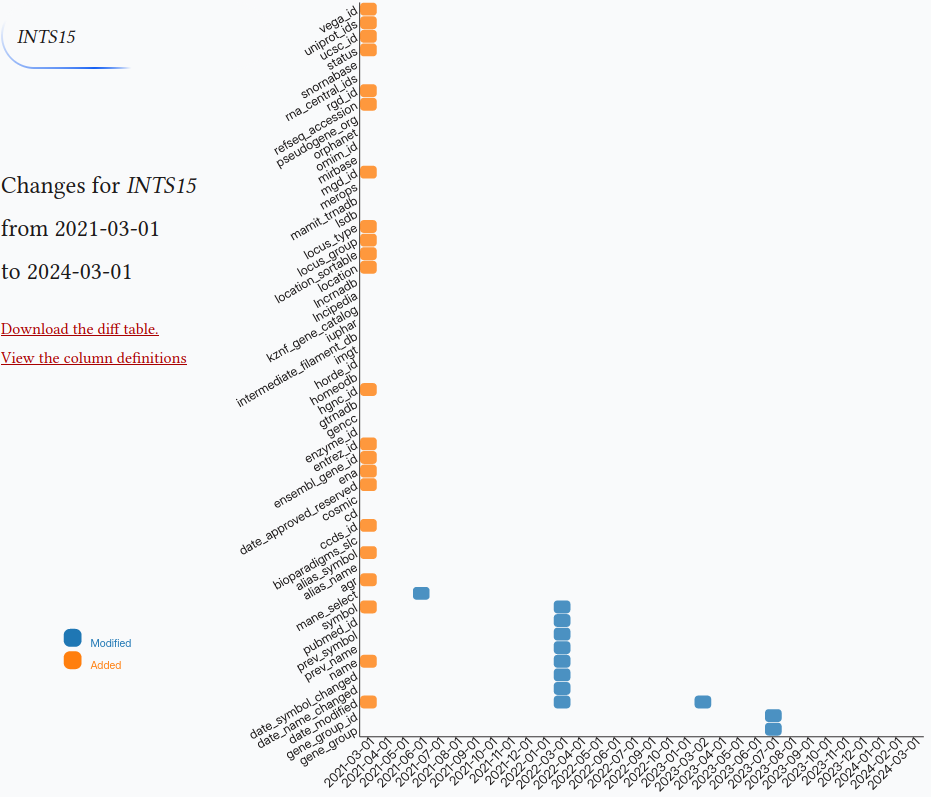
This tool shows the differences of gene symbols over the months on a heatmap. HGNC archive tables have been flattened out to the files and have been committed to a git repo in a chronological order. The data repo can be found at github.
Cross-symbol checker

This tool maps aliases, and previous or withdrawn symbols to current approved HGNC symbols as well as check for any entries that are not gene symbols and fix capitalization. Ensembl and RefSeq annotation files sometimes use previous or alias symbols. This tool also cross checks against Ensembl and NCBI annotation files for different genome versions and shows which gene symbols used. This way an accurate gene set can be utilized to avoid false negatives in the variant discovery process. The source code is available on GitHub.